Identification, classification, and prioritization of most influential players in normal biological processes and diseases
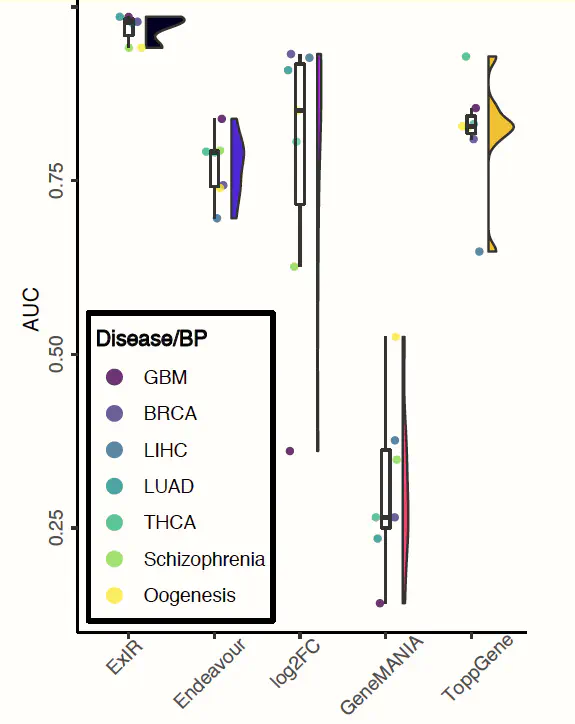 Image credit: Adrian Salavaty
Image credit: Adrian SalavatyAbstract
Current sequencing technologies have enabled the identification and measuring of the activity of thousands of genes and proteins at a time and across several conditions and cell types. This has a huge benefit for studying the basis as well as the drivers of cancer progression. However, one of the biggest challenges is the selection of right candidates amongst thousands of features for experimental functional validation. Currently, a number of different models have been developed for candidate gene prioritization, most of which rely on external sources of information and, consequently, are not applicable to all data types. Also, most of the available general-purpose models are not optimized for handling and analyzing single-cell data. Moreover, current models are not able to classify genes into “drivers”, “biomarkers”, and “mediators” according to their functional importance. Here we present, ExIR—Experimental data-based Integrative Ranking— which recruits the potential of Integrated Value of Influence (IVI) algorithm and combines network reconstruction and machine learning techniques to extract, classify and prioritize candidate features from any type of experimental data such as bulk and single-cell RNA sequencing. The evaluation of ExIR in the context of real-world experimental data confirmed its superiority to other respective contemporary methods and algorithms.
Key words
IVI value; influential node; experimental-data-based integrative ranking; ExIR; gene classification; feature prioritization; network analysis; machine learning; systems biology